pyanno Package¶
Other modules¶
abstract_model Module¶
Defines AbstractModel, an abstract class that specifies the interface of pyAnno models.
- class pyanno.abstract_model.AbstractModel[source]¶
Bases: traits.has_traits.HasTraits
Abstract class defining the interface of a pyAnno model.
- are_annotations_compatible(annotations)[source]¶
Returns True if the annotations are compatible with the model.
The standard implementation is: valid if the number of annotators is correct, if the classes are between 0 and nclasses-1, and if missing values are marked with pyanno.util.MISSING_VALUE
- static create_initial_state(nclasses)[source]¶
Factory method returning a model with random initial parameters.
Parameters: nclasses (int) – Number of label classes
- generate_annotations(nitems)[source]¶
Generate a random annotation set from the model.
Sample a random set of annotations from the probability distribution defined the current model parameters.
Parameters: nitems (int) – Number of items to sample Returns: annotations (ndarray, shape = (n_items, n_annotators)) - annotations[i,j] is the annotation of annotator j for item i
- infer_labels(annotations)[source]¶
Infer posterior distribution over label classes.
Compute the posterior distribution over label classes given observed annotations,
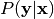 .
.- annotations : ndarray, shape = (n_items, n_annotators)
- annotations[i,j] is the annotation of annotator j for item i
Returns: posterior (ndarray, shape = (n_items, n_classes)) - posterior[i,k] is the posterior probability of class k given the annotation observed in item i.
- log_likelihood(annotations)[source]¶
Compute the log likelihood of a set of annotations given the model.
Returns log P(annotations | current model parameters).
Parameters: annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation of annotator j for item i Returns: log_lhood (float) - log likelihood of annotations
- map(annotations)[source]¶
Computes maximum a posteriori (MAP) estimate of parameters.
Estimate the model parameters from a set of observed annotations using maximum a posteriori estimation.
Parameters: annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation of annotator j for item i
- mle(annotations)[source]¶
Computes maximum likelihood estimate (MLE) of parameters.
Estimate the model parameters from a set of observed annotations using maximum likelihood estimation.
Parameters: annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation of annotator j for item i
- sample_posterior_over_accuracy(annotations, nsamples, burn_in_samples=0, thin_samples=1)[source]¶
Return samples from posterior over the accuracy parameters.
Draw samples from P(accuracy parameters | data, model parameters). The accuracy parameters control the probability of an annotator reporting the correct label (the exact nature of these parameters varies from model to model).
Parameters: - annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation of annotator j for item i
- nsamples (int) – Number of samples to return (i.e., burn-in and thinning samples are not included)
- burn_in_samples (int) – Discard the first burn_in_samples during the initial burn-in phase, where the Monte Carlo chain converges to the posterior
- thin_samples (int) – Only return one every thin_samples samples in order to reduce the auto-correlation in the sampling chain. This is called “thinning” in MCMC parlance.
Returns: samples (ndarray, shape = (n_samples, ??)) - Array of samples from the posterior distribution over parameters.
util Module¶
Utility functions.
- exception pyanno.util.PyannoValueError[source]¶
Bases: exceptions.ValueError
ValueError subclass raised by pyAnno functions and methods.
- class pyanno.util.benchmark(name)[source]¶
Bases: object
Simple context manager to simplify benchmarking.
Usage:
with benchmark('fast computation'): do_something()
- pyanno.util.compute_counts(annotations, nclasses)[source]¶
Transform annotation data in counts format.
At the moment, it is hard coded for 8 annotators, 3 annotators active at any time.
Parameters: - annotations (ndarray, shape = (n_items, 8)) – Annotations array
- nclasses (int) – Numer of label classes
Returns: data (ndarray, shape = (n_classes^3, 9)) - data[i,m] is the number of times the combination of annotators number m voted according to pattern i
- pyanno.util.create_band_matrix(shape, diagonal_elements)[source]¶
Create a symmetrical band matrix from a list of elements.
Parameters: - shape (int) – Width of the matrix
- diagonal_elements (list or array) – List of elements in the first row. If the list is smaller than shape, the last element is used to fill the the remaining items.
- pyanno.util.dirichlet_llhood(theta, alpha)[source]¶
Compute the log likelihood of theta under Dirichlet(alpha).
Parameters: - theta (ndarray) – Categorical probability distribution. theta[i] is the probability of item i. Elements of the array have to sum to 1.0 (not forced for efficiency reasons)
- alpha (ndarray) – Parameters of the Dirichlet distribution
Returns: log_likelihood (float) - Log lihelihood of theta given alpha
- pyanno.util.is_valid(annotations)[source]¶
Return True if annotation is valid.
An annotation is valid if it is not equal to the missing value, MISSING_VALUE.
- pyanno.util.labels_count(annotations, nclasses, missing_val=-1)[source]¶
Compute the total count of labels in observed annotations.
Parameters: - annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation made by annotator j on item i
- nclasses (int) – Number of label classes in annotations
- missing_val (int) – Value used to indicate missing values in the annotations. Default is MISSING_VALUE
Returns: count (ndarray, shape = (n_classes, )) - count[k] is the number of elements of class k in annotations
- pyanno.util.labels_frequency(annotations, nclasses, missing_val=-1)[source]¶
Compute the total frequency of labels in observed annotations.
Parameters: - annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation made by annotator j on item i
- nclasses (int) – Number of label classes in annotations
- missing_val (int) – Value used to indicate missing values in the annotations. Default is MISSING_VALUE
Returns: freq (ndarray, shape = (n_classes, )) - freq[k] is the frequency of elements of class k in annotations, i.e. their count over the number of total of observed (non-missing) elements
- pyanno.util.majority_vote(annotations)[source]¶
Compute an estimate of the real class by majority vote.
In case of ties, return the class with smallest number.
Parameters: annotations (ndarray, shape = (n_items, n_annotators)) – annotations[i,j] is the annotation made by annotator j on item i - vote : ndarray, shape = (n_items, )
- vote[i] is the majority vote estimate for item i
- pyanno.util.normalize(x, dtype=<type 'float'>)[source]¶
Returns a normalized distribution (sums to 1.0).
If x consists only of zero element, the returned array has elements 1/n , where n is the length of x.
- pyanno.util.random_categorical(distr, nsamples)[source]¶
Return an array of samples from a categorical distribution.
Parameters: - distr (ndarray) – distr[i] is the probability of item i
- nsamples (int) – Number of samples to draw from the distribution
Returns: samples (ndarray, shape = (n_samples, )) - Samples from the distribution
- pyanno.util.MISSING_VALUE = -1¶
In annotations arrays, this is the value used to indicate missing values
- pyanno.util.SMALLEST_FLOAT = -1.7976931348623157e+308¶
Smallest possible floating point number, somtimes used instead of -np.inf to make numberical calculation return a meaningful value
sampling Module¶
This module defines functions to sample from a distribution given its log likelihood.
- pyanno.sampling.optimize_step_size(likelihood, x0, arguments, x_lower, x_upper, n_samples, recomputing_cycle, target_rejection_rate, tolerance)[source]¶
Compute optimum jump for MCMC estimation of credible intervals.
Adjust jump size in Metropolis-Hasting MC to achieve target rejection rate. Jump size is estimated for each parameter separately.
Parameters: - likelihood (function(params, arguments)) – Function returning the unnormalized log likelihood of data given the parameters. The function accepts two arguments: params is the vector of parameters; arguments contains any additional argument that is needed to compute the log likelihood.
- x0 (ndarray, shape = (n_parameters, )) – Initial parameters value.
- arguments (any) – Additional argument passed to the function likelihood.
- x_lower (ndarray, shape = (n_parameters, )) – Lower bound for the parameters.
- x_upper (ndarray, shape = (n_parameters, )) – Upper bound for the parameters.
- n_samples (int) – Total number of samples to draw during the optimization.
- recomputing_cycle (int) – Number of samples over which the rejection rates are computed. After recomputing_cycle samples, the step size is adapted and the rejection rates are reset to 0.
- target_rejection_rate (float) – Target rejection rate. If the rejection rate over the latest cycle is closer than tolerance to this target, the optimization phase is concluded.
- tolerance (float) – Tolerated deviation from target_rejection_rate.
Returns: step (ndarray, shape = (n_parameters, )) - The final optimized step size.
- pyanno.sampling.sample_distribution(likelihood, x0, arguments, step, nsamples, x_lower, x_upper)[source]¶
General-purpose sampling routine for MCMC sampling.
Draw samples from a distribution given its unnormalized log likelihood using the Metropolis-Hasting Monte Carlo algorithm.
It is recommended to optimize the step size, step, using the function optimize_step_size() in order to reduce the autocorrelation between successive samples.
Parameters: - likelihood (function(params, arguments)) – Function returning the unnormalized log likelihood of data given the parameters. The function accepts two arguments: params is the vector of parameters; arguments contains any additional argument that is needed to compute the log likelihood.
- x0 (ndarray, shape = (n_parameters, )) – Initial parameters value.
- arguments (any) – Additional argument passed to the function likelihood.
- step (ndarray, shape = (n_parameters, )) – Width of the proposal distribution over the parameters.
- nsamples (int) – Number of samples to draw from the distribution.
- x_lower (ndarray, shape = (n_parameters, )) – Lower bound for the parameters.
- x_upper (ndarray, shape = (n_parameters, )) – Upper bound for the parameters.
- pyanno.sampling.sample_from_proposal_distribution(theta, step, lower, upper)[source]¶
Returns one sample from the proposal distribution.
Parameters: - theta (float) – current parameter value
- step (float) – width of the proposal distribution over theta
- lower (float) – lower bound for theta
- upper (float) – upper bound for theta
Returns: - theta_new (float) - new sample from the distribution over theta
- log_q_ratio (float) - log-ratio of probability of new value given old value to probability of old value given new value